
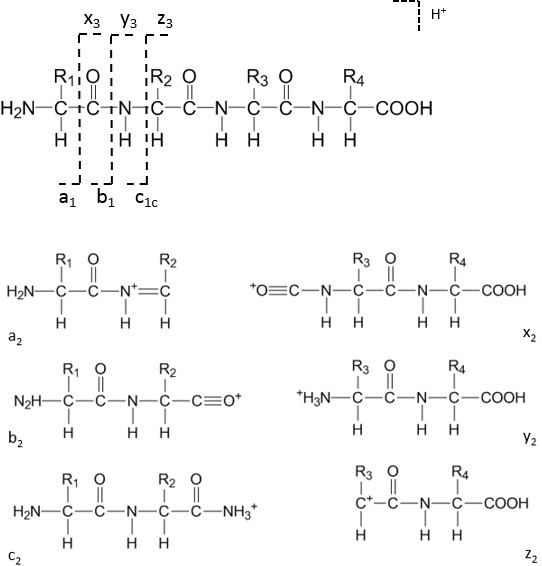
The “depth of coverage” for an amino acid is defined as the number of unique PSMs covering the amino acid. The second and third columns list the median depths of coverage of the HCDR3 and the other variable portions, respectively. The first column lists the protease combination, where each protease is represented by a single letter code: P (Pepsin), T (Trypsin), C (Chymotrypsin), A (Asp-N), and L (Lys-C). The median depth of coverage for the HCDR3 region and the remaining portion of the variable regions are listed in the following table for each combination of the proteases used in this study. Sequence motif analysis was performed using SeqtoLogo. Yeast complex samples were analyzed with Search GUI-3.3.13 and PeptideShaker-1.16.37 using no enzyme restriction, 25ppm with protein, peptide and 1% psm FDR, while considering a combination of X! tandem, MS-GF+ and Comet. Data analysis:ĭe novo peptide sequencing was performed with Novor search algorithm and the protein sequences were assembled with REmAb ® ( Figure 1). ETD spectra were acquired with three different collisions energies.
#DE NOVO PROTEIN SEQUENCE ANALYSIS SERIES#
Protein lysates were analyzed with an Orbitrap Fusion™ Series Tribid™ instrument (ThermoFisher Scientific, CA, US) coupled to the LC Evosep One (Evosep, Denmark) in both HCD and ETD mode. department of Commerce), monoclonal mAb04 and yeast extracts (Promega, WI, US). Trypsin, Lys-C, Chymotrypsin, Pepsin, A/P, and recombinant Asp-N proteases Promega (Promega, WI, US) were used to digest NISTmAb humanized IgG1 monoclonal antibody RM8671 (National Institute of Standards and technology, U.S.
#DE NOVO PROTEIN SEQUENCE ANALYSIS FULL#


 0 kommentar(er)
0 kommentar(er)
